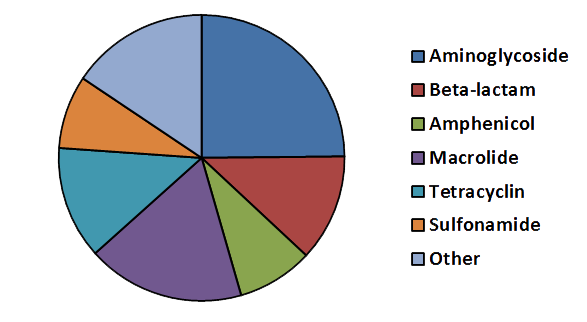The resqu database
Introduction
Antibiotic resistance is caused by changes in the bacterial genome. These changes are often caused by the acquisition of antibiotic resistance genes, which are mobile and can rapidly spread between bacteria using the process of horizontal transfer. Almost all forms of antibiotics are today associated with resistance genes and several thousands of variants have been described by the scientific literature.
The resqu database
The resqu database collects horizontally transferred antibiotic resistance genes. Each gene, together with its DNA sequences, the encoded protein, and a large set of metadata is organized in a structure describing its resistance mechanisms and genetic properties. The metadata contains information about its host, range of mobility, biochemical properties and induced resistance phenotype. In contrast to most databases for antibiotic resistance genes, all data in resqu is manually extracted from the scientific literature and public sequence repositories. All sequences and metadata is also quality assessed and manually curated. In particularly, the database contains only genes where
- The resistance phenotype for each gene have been experimentally verified,
- There is ample evidence for horizontal transfer between at least two different species,
- The DNA/protein sequences have been manually assessed for errors (e.g. sequencing errors, start and stop of the open reading frame) and are non-redundant,
- The DNA/protein sequences show a consistency within established gene families (manually assessed by multiple alignments).
Current version
The current version of resqu (version 1.1) contains 325 families of resistance genes described by 3018 non-redundant DNA and protein sequences. These sequences are distributed according to their resistance mechanisms as described by the pie chart to the right.
Contact
The resqu resistance gene database is developed by 1928 Diagnostics AB (Gothenburg, Sweden) in collaboration with Chalmers University of Technology (Gothengurg, Sweden). For inquiries about the resqu resistance database please contact erik.kristiansson@1928d.com.